Project 2: Regulation of FLOWERING LOCUS T by distal enhancers and epigenetic repressors
Among functionally related gene groups highly enriched for PcG targets are those that promote reproductive development. Deciding when to flower is one of the most crucial decisions in a plant’s life cycle and therefore requires integration of environmental and developmental signals into a complex gene regulatory network. PcG targets promoting flowering are transcriptionally repressed during vegetative development while others, that repress flowering, acquire a PcG-repressed state in response to environmental or developmental cues. We have selected FLOWERING LOCUS T (FT) for a detailed study of transcriptional regulation. FT and FT-like genes encode mobile proteins that act as a flowering hormone in many plant species. We showed that FT transcriptional regulation in response to photoperiod in leaves is dependent on two distal enhancers representing accessible chromatin islands within PcG-repressed chromatin [1,2,3]. Current and future efforts aim to elucidate how distinct distal enhancers can dynamically shift their contribution to FT expression dependent on tissue-type and the perception of different external cues.
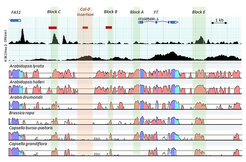
Chromatin accessibility, repressive H3K27me3 profile and sequence conservation level at the FT locus pinpoint distal enhancers. Chromatin accessibility at the FT locus as determined by DNase-seq data (labelled DNase I). A measure of chromatin compactness is the presence of the epigenetic histone modification H3K27me3 (labelled as H3K27me3). The tracks below show the level of sequence homology between A. thaliana and six related Brassicaceae species. Red, cyan and purple colours code indicate intergenic regions and introns, Untranslated Regions (UTRs) and exons, respectively. Green shadows indicate regions Block C, Block B, and Block E, which show both, conserved sequence and local open chromatin. The FT promoter, Block A, is in a closed chromatin conformation, which requires the presence of Block C and Block E for FT transcriptional induction in response to long day photoperiod. The orange shadow indicates the location of a large insertion/deletion polymorphism within the A. thaliana species.
[1] Adrian, J., S. Farrona, J.J. Reimer, M.C. Albani, G. Coupland and F. Turck (2010): cis-Regulatory Elements and Chromatin State Coordinately Control Temporal and Spatial Expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22, 1425-1440 (2010). open access
[2] Liu L., Adrian J., Pankin A., Hu J., Dong X., von Korff M., and F. Turck. Induced and natural variation of promoter length modulates the photoperiodic response of FLOWERING LOCUS T. Nat Commun. 5:4558, doi: 10.1038/ncomms5558 (2014). Open access
[3] Zicola, J., Liu, L., Tänzler, P., Turck, F. (2019) Targeted DNA methylation represses two enhancers of FLOWERING LOCUS T in Arabidopsis thaliana. Nature Plants 5, 300–307 doi: 10.1038/s41477-019-0375-2 . open access
